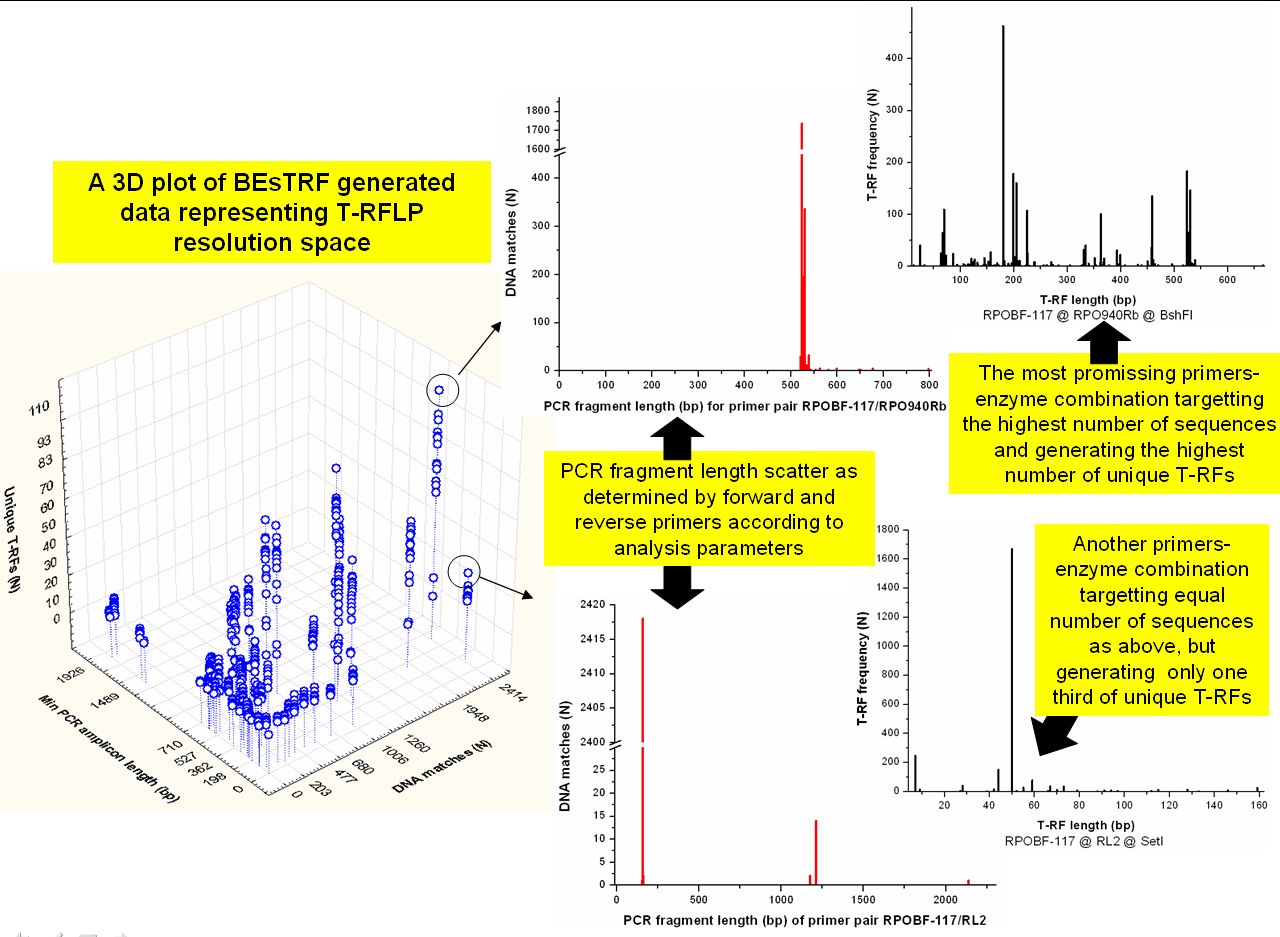
Click to enlarge
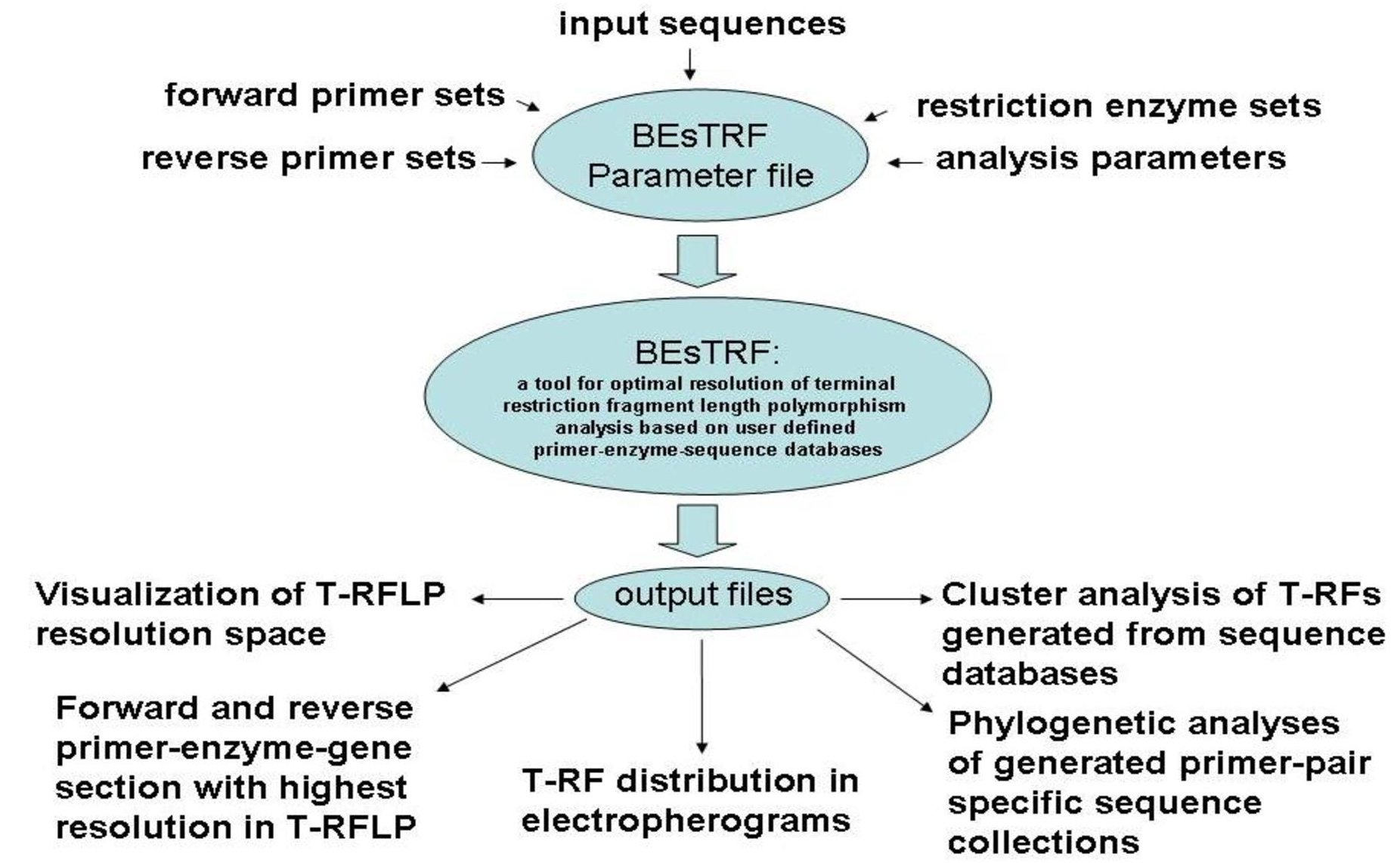
BEsTRF: a tool for optimal resolution of terminal
restriction fragment length polymorphism analysis based on user
defined primer-enzyme-sequence databases
|
||
Reference for citation: |
||
BEsTRF: a tool for optimal resolution of terminal-restriction fragment
length polymorphism (T-RFLP) analysis based on user defined primer-enzyme-sequence databases
|
||
Free access link to abstract: http://bioinformatics.oxfordjournals.org/cgi/content/abstract/btp254?ijkey=qw9y7y9LEcKhXd7&keytype=ref |
||

|
|